4 Data Joins and Transformations
Reading: 17 minute(s) at 200 WPM.
Videos: 26 minutes.
Objectives
The first section of this chapter is heavily outsourced to r4ds as they do a much better job at providing examples and covering the extensive functionality of factor data types than I myself would ever be able to.
- Use
forcatsto reorder and relabel factor variables in data cleaning steps and data visualizations.
Broadly, your objective while reading the second section of this chapter is to be able to identify data sets which have “messy” formats and determine a sequence of operations to transition the data into “tidy” format. To do this, you should master the following concepts:
- Determine what data format is necessary to generate a desired plot or statistical model.
- Understand the differences between “wide” and “long” format data and how to transition between the two structures.
- Understand relational data formats and how to use data joins to assemble data from multiple tables into a single table.
Functions covered this week:
library(forcats) (not an exhaustive list)
Download the forcats cheatsheet.
-
pivot_longer(),pivot_wider() -
separate(),unite()
-
left_join(),right_join(),full_join() -
semi_join(),anti_join()
4.1 Factors with forcats
We have been floating around the idea of factor data types. In this section, we will formally define factors and why they are needed for data visualization and analysis. We will then learn useful functions for working with factors in our data cleaning steps.
In short, factors are categorical variables with a fixed number of values (think a set number of groups). One of the main features that set factors apart from groups is that you can reorder the groups to be non-alphabetical. In this section we will be using the forcats package (part of the tidyverse!) to create and manipulate factor variables.
(Required) Go read about factors in r4ds.

4.2 Identifying the problem: Messy data
The illustrations below are lifted from an excellent blog post (Lowndes and Horst 2020) about tidy data; they’re reproduced here because
- they’re beautiful and licensed as CCA-4.0-by, and
- they might be more memorable than the equivalent paragraphs of text without illustration.
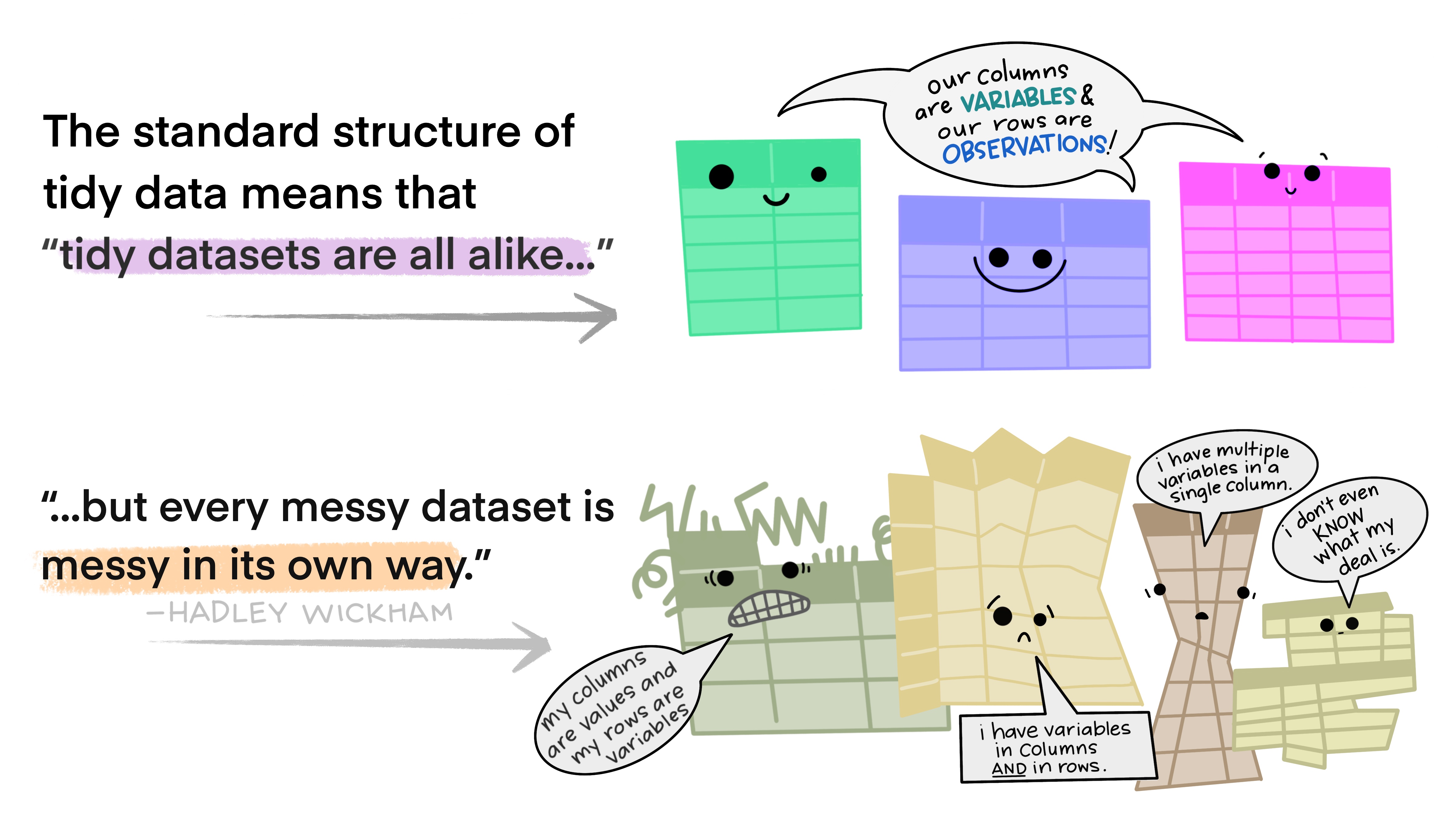
Most of the time, data does not come in a format suitable for analysis. Spreadsheets are generally optimized for data entry or viewing, rather than for statistical analysis:
- Tables may be laid out for easy data entry, so that there are multiple observations in a single row
- It may be visually preferable to arrange columns of data to show multiple times or categories on the same row for easy comparison
When we analyze data, however, we care much more about the fundamental structure of observations: discrete units of data collection. Each observation may have several corresponding variables that may be measured simultaneously, but fundamentally each discrete data point is what we are interested in analyzing or plotting.
The structure of tidy data reflects this preference for keeping the data in a fundamental form: each observation is in its own row, any observed variables are in single columns. This format is inherently rectangular, which is also important for statistical analysis - our methods are typically designed to work with matrices of data.
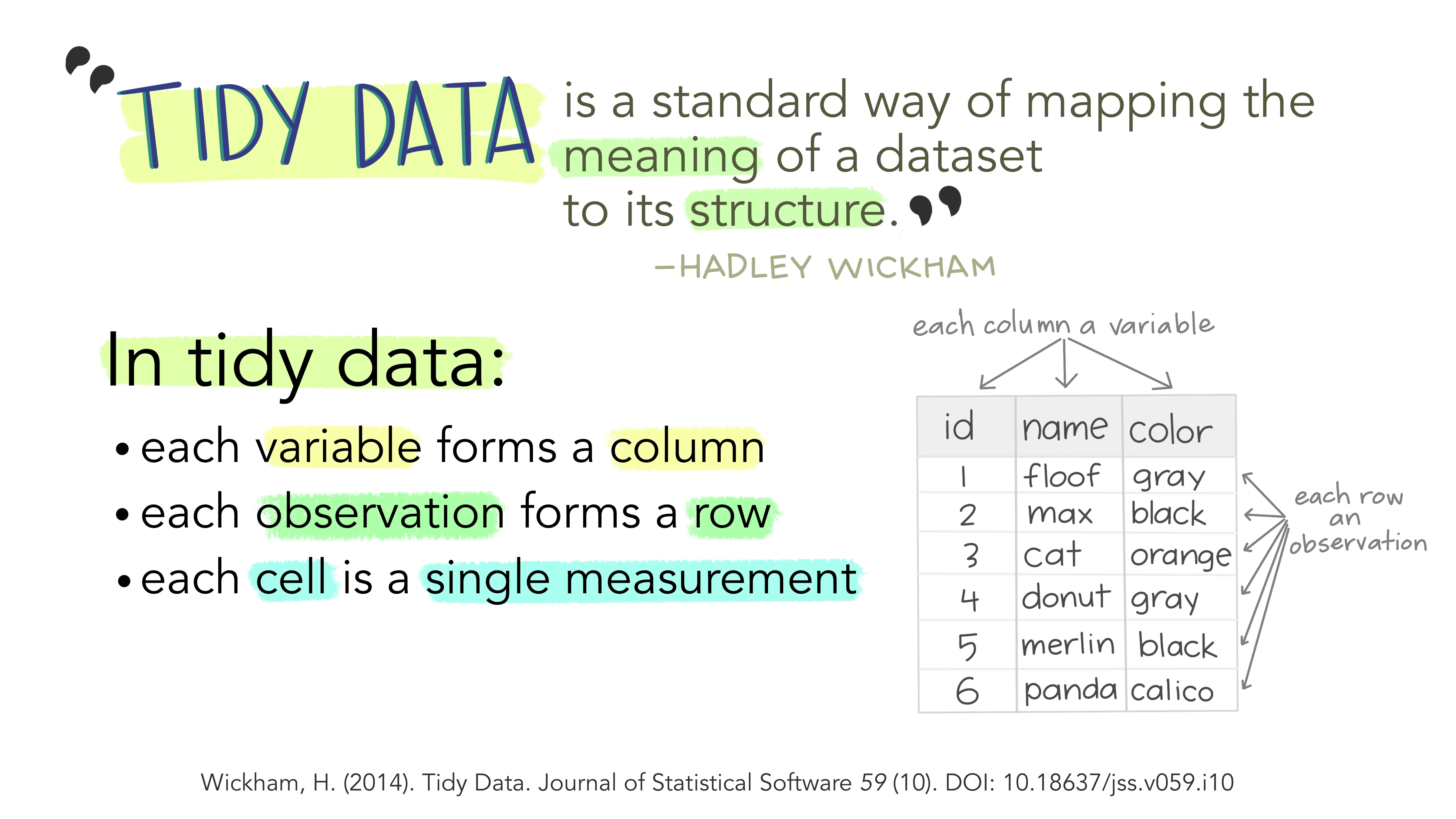
The preference for tidy data has several practical implications: it is easier to reuse code on tidy data, allowing for analysis using a standardized set of tools (rather than having to build a custom tool for each data analysis job).
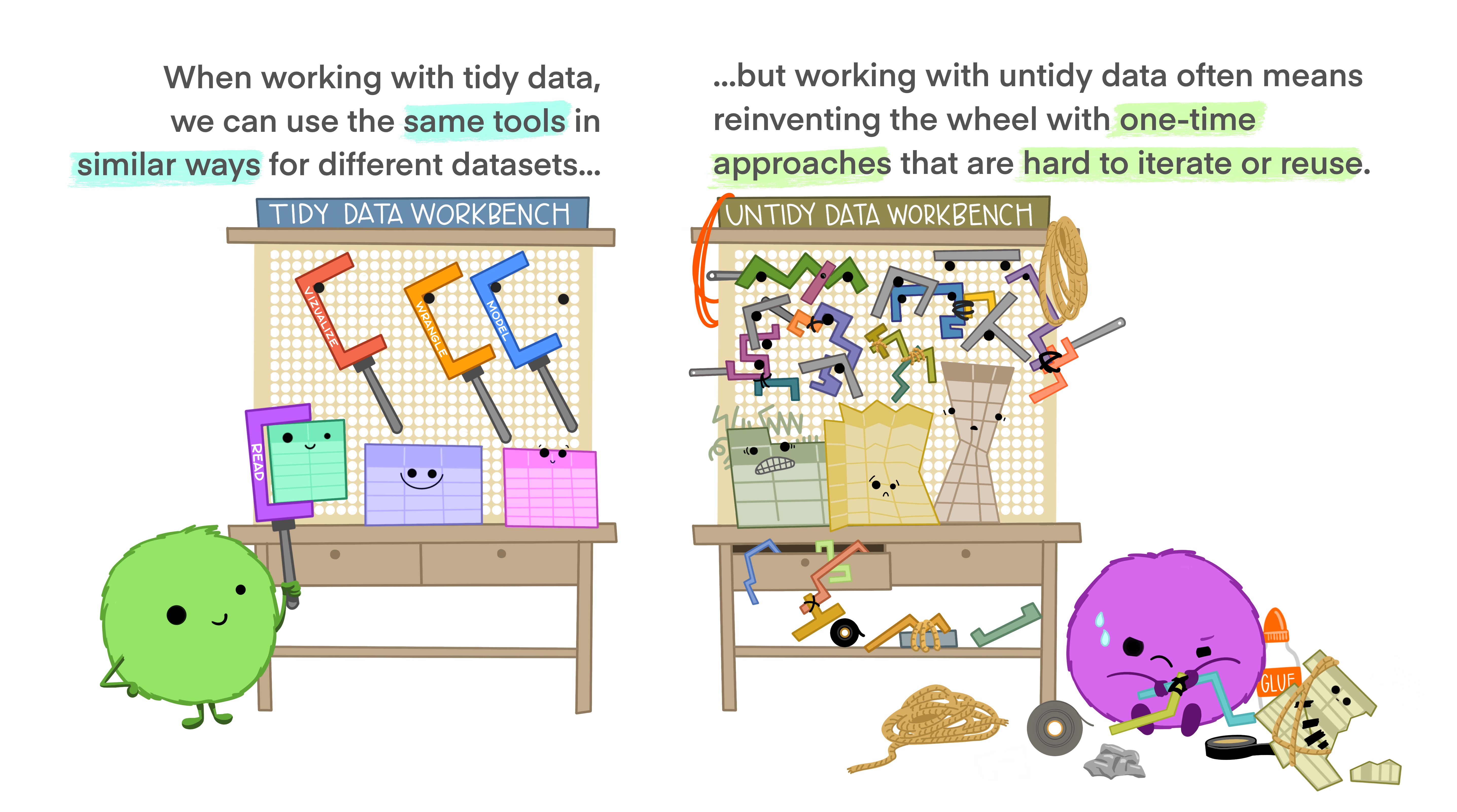
In addition, standardized tools for data analysis means that it is easier to collaborate with others: if everyone starts with the same set of assumptions about the dataset, you can borrow methods and tools from a collaborator’s analysis and easily apply them to your own dataset.
Examples: Messy Data
These datasets all display the same data: TB (Tuberculosis) cases documented by the WHO (World Health Organization) in Afghanistan, Brazil, and China, between 1999 and 2000. There are 4 variables: country, year, cases, and population, but each table has a different layout.
For each of the data set, determine whether each table is tidy. If it is not, identify which rule or rules it violates.
What would you have to do in order to compute a standardized TB infection rate per 100,000 people?
All of these data sets are “built-in” to the tidyverse package
| country | year | cases | population |
|---|---|---|---|
| Afghanistan | 1999 | 745 | 19987071 |
| Afghanistan | 2000 | 2666 | 20595360 |
| Brazil | 1999 | 37737 | 172006362 |
| Brazil | 2000 | 80488 | 174504898 |
| China | 1999 | 212258 | 1272915272 |
| China | 2000 | 213766 | 1280428583 |
Here, each observation is a single row, each variable is a column, and everything is nicely arranged for e.g. regression or statistical analysis. We can easily compute another measure, such as cases per 100,000 population, by taking cases/population * 100000 (this would define a new column).
| country | year | type | count |
|---|---|---|---|
| Afghanistan | 1999 | cases | 745 |
| Afghanistan | 1999 | population | 19987071 |
| Afghanistan | 2000 | cases | 2666 |
| Afghanistan | 2000 | population | 20595360 |
| Brazil | 1999 | cases | 37737 |
| Brazil | 1999 | population | 172006362 |
| Brazil | 2000 | cases | 80488 |
| Brazil | 2000 | population | 174504898 |
| China | 1999 | cases | 212258 |
| China | 1999 | population | 1272915272 |
| China | 2000 | cases | 213766 |
| China | 2000 | population | 1280428583 |
Here, we have 4 columns again, but we now have 12 rows: one of the columns is an indicator of which of two numerical observations is recorded in that row; a second column stores the value. This form of the data is more easily plotted in e.g. ggplot2, if we want to show lines for both cases and population, but computing per capita cases would be much more difficult in this form than in the arrangement in table 1.
| country | year | rate |
|---|---|---|
| Afghanistan | 1999 | 745/19987071 |
| Afghanistan | 2000 | 2666/20595360 |
| Brazil | 1999 | 37737/172006362 |
| Brazil | 2000 | 80488/174504898 |
| China | 1999 | 212258/1272915272 |
| China | 2000 | 213766/1280428583 |
This form has only 3 columns, because the rate variable (which is a character) stores both the case count and the population. We can’t do anything with this format as it stands, because we can’t do math on data stored as characters. However, this form might be easier to read and record for a human being.
| country | 1999 | 2000 |
|---|---|---|
| Afghanistan | 745 | 2666 |
| Brazil | 37737 | 80488 |
| China | 212258 | 213766 |
| country | 1999 | 2000 |
|---|---|---|
| Afghanistan | 19987071 | 20595360 |
| Brazil | 172006362 | 174504898 |
| China | 1272915272 | 1280428583 |
In this form, we have two tables - one for population, and one for cases. Each year’s observations are in a separate column. This format is often found in separate sheets of an excel workbook. To work with this data, we’ll need to transform each table so that there is a column indicating which year an observation is from, and then merge the two tables together by country and year.
| country | century | year | rate |
|---|---|---|---|
| Afghanistan | 19 | 99 | 745/19987071 |
| Afghanistan | 20 | 00 | 2666/20595360 |
| Brazil | 19 | 99 | 37737/172006362 |
| Brazil | 20 | 00 | 80488/174504898 |
| China | 19 | 99 | 212258/1272915272 |
| China | 20 | 00 | 213766/1280428583 |
Table 5 is very similar to table 3, but the year has been separated into two columns - century, and year. This is more common with year, month, and day in separate columns (or date and time in separate columns), often to deal with the fact that spreadsheets don’t always handle dates the way you’d hope they would.
By the end of this chapter, you will have the skills needed to wrangle and transform the most common “messy” data sets into “tidy” form.
4.3 Pivot Operations
It’s fairly common for data to come in forms which are convenient for either human viewing or data entry. Unfortunately, these forms aren’t necessarily the most friendly for analysis.

The two operations we’ll learn here are wide -> long and long -> wide.

This animation uses the functions pivot_wider() and pivot_longer() from the tidyr package in R – Animation source.
4.3.1 Longer
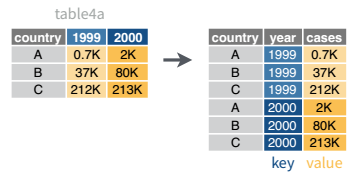
In many cases, the data come in what we might call “wide” form - some of the column names are not names of variables, but instead, are themselves values of another variable.
Tables 4a and 4b (from above) are good examples of data which is in “wide” form and should be in long(er) form: the years, which are variables, are column names, and the values are cases and population respectively.
table4a# A tibble: 3 × 3
country `1999` `2000`
<chr> <dbl> <dbl>
1 Afghanistan 745 2666
2 Brazil 37737 80488
3 China 212258 213766table4b# A tibble: 3 × 3
country `1999` `2000`
<chr> <dbl> <dbl>
1 Afghanistan 19987071 20595360
2 Brazil 172006362 174504898
3 China 1272915272 1280428583The solution to this is to rearrange the data into “long form”: to take the columns which contain values and “stack” them, adding a variable to indicate which column each value came from. To do this, we have to duplicate the values in any column which isn’t being stacked (e.g. country, in both the example above and the image below).
Once our data are in long form, we can (if necessary) separate values that once served as column labels into actual variables, and we’ll have tidy(er) data.
table4a |>
pivot_longer(cols = `1999`:`2000`,
names_to = "year",
values_to = "cases"
)# A tibble: 6 × 3
country year cases
<chr> <chr> <dbl>
1 Afghanistan 1999 745
2 Afghanistan 2000 2666
3 Brazil 1999 37737
4 Brazil 2000 80488
5 China 1999 212258
6 China 2000 213766table4b |>
pivot_longer(cols = -country,
names_to = "year",
values_to = "population"
)# A tibble: 6 × 3
country year population
<chr> <chr> <dbl>
1 Afghanistan 1999 19987071
2 Afghanistan 2000 20595360
3 Brazil 1999 172006362
4 Brazil 2000 174504898
5 China 1999 1272915272
6 China 2000 1280428583The columns are moved to a variable with the name passed to the argument “names_to” (hopefully, that is easy to remember), and the values are moved to a variable with the name passed to the argument “values_to” (again, hopefully easy to remember).
We identify ID variables (variables which we don’t want to pivot) by not including them in the pivot statement. We can do this in one of two ways:
- select only variables (columns) we want to pivot (see table4a pivot)
- select variables (columns) we don’t want to pivot, using
-to remove them (see table4b pivot)
Which option is easier depends how many things you’re pivoting (and how the columns are structured).
4.3.2 Wider
While it’s very common to need to transform data into a longer format, it’s not that uncommon to need to do the reverse operation. When an observation is scattered across multiple rows, your data is too long and needs to be made wider again.
Table 2 (from above) is an example of a table that is in long format but needs to be converted to a wider layout to be “tidy” - there are separate rows for cases and population, which means that a single observation (one year, one country) has two rows.
table2# A tibble: 12 × 4
country year type count
<chr> <dbl> <chr> <dbl>
1 Afghanistan 1999 cases 745
2 Afghanistan 1999 population 19987071
3 Afghanistan 2000 cases 2666
4 Afghanistan 2000 population 20595360
5 Brazil 1999 cases 37737
6 Brazil 1999 population 172006362
7 Brazil 2000 cases 80488
8 Brazil 2000 population 174504898
9 China 1999 cases 212258
10 China 1999 population 1272915272
11 China 2000 cases 213766
12 China 2000 population 1280428583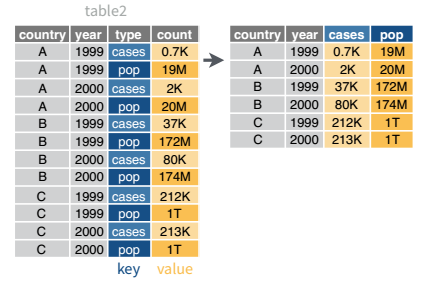
table2 |>
pivot_wider(names_from = type,
values_from = count
)# A tibble: 6 × 4
country year cases population
<chr> <dbl> <dbl> <dbl>
1 Afghanistan 1999 745 19987071
2 Afghanistan 2000 2666 20595360
3 Brazil 1999 37737 172006362
4 Brazil 2000 80488 174504898
5 China 1999 212258 1272915272
6 China 2000 213766 1280428583Learn More in R4DS
Read more about pivoting in r4ds.
4.4 Separating and Uniting Variables
We will talk about strings and regular expressions next week, but there’s a task that is fairly commonly encountered with functions that belong to the tidyr package: separating variables into two different columns separate() and it’s complement, unite(), which is useful for combining two variables into one column.
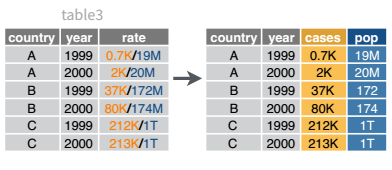
table3 |>
separate_wider_delim(cols = rate,
names = c("cases", "population"),
delim = "/",
cols_remove = FALSE
)# A tibble: 6 × 5
country year cases population rate
<chr> <dbl> <chr> <chr> <chr>
1 Afghanistan 1999 745 19987071 745/19987071
2 Afghanistan 2000 2666 20595360 2666/20595360
3 Brazil 1999 37737 172006362 37737/172006362
4 Brazil 2000 80488 174504898 80488/174504898
5 China 1999 212258 1272915272 212258/1272915272
6 China 2000 213766 1280428583 213766/1280428583I’ve left the rate column in the original data frame (cols_remove = F) just to make it easy to compare and verify that yes, it worked.
And, of course, there is a complementary operation, which is when it’s necessary to join two columns to get a useable data value.
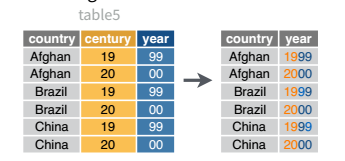
table5 |>
unite(col = "year",
c(century, year),
sep = ''
)# A tibble: 6 × 3
country year rate
<chr> <chr> <chr>
1 Afghanistan 1999 745/19987071
2 Afghanistan 2000 2666/20595360
3 Brazil 1999 37737/172006362
4 Brazil 2000 80488/174504898
5 China 1999 212258/1272915272
6 China 2000 213766/1280428583Learn More in R4DS
The separate_xxx() is actually a family of experimental functions stemming from the superseeded separate() function. You can read more about separate_xxx() and unite() in r4ds and r4ds.
4.5 Merging Tables
The final essential data tidying and transformation skill you need to acquire is joining tables. It is common for data to be organized relationally - that is, certain aspects of the data apply to a group of data points, and certain aspects apply to individual data points, and there are relationships between the individual data points and the groups of data points that have to be documented.
Examples: Relational Data Example: Primary School Records
Each individual has certain characteristics:
- full_name
- gender
- birth date
- ID number
Each student has specific characteristics:
- ID number
- parent name
- parent phone number
- medical information
- Class ID
Teachers may also have additional information:
- ID number
- Class ID
- employment start date
- education level
- compensation level
There are also fields like grades, which occur for each student in each class, but multiple times a year.
- ID number
- Student ID
- Class ID
- year
- term number
- subject
- grade
- comment
And for teachers, there are employment records on a yearly basis
- ID number
- Employee ID
- year
- rating
- comment
But each class also has characteristics that describe the whole class as a unit:
- location ID
- class ID
- meeting time
- grade level
Each location might also have some logistical information attached:
- location ID
- room number
- building
- number of seats
- AV equipment
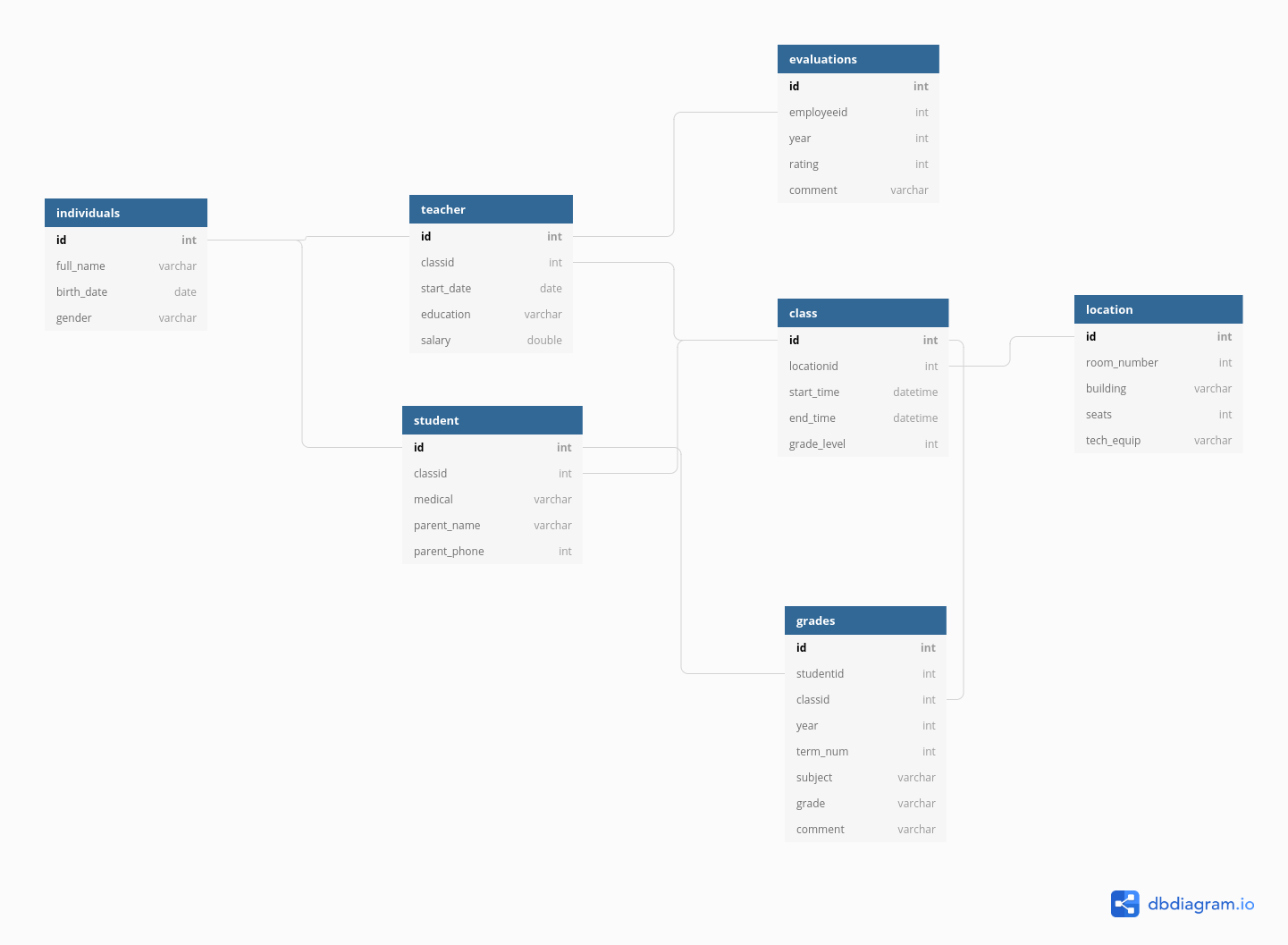
We could go on, but you can see that this data is hierarchical, but also relational: - each class has both a teacher and a set of students - each class is held in a specific location that has certain equipment
It would be silly to store this information in a single table (though it probably can be done) because all of the teacher information would be duplicated for each student in each class; all of the student’s individual info would be duplicated for each grade. There would be a lot of wasted storage space and the tables would be much more confusing as well.
But, relational data also means we have to put in some work when we have a question that requires information from multiple tables. Suppose we want a list of all of the birthdays in a certain class. We would need to take the following steps:
- get the Class ID
- get any teachers that are assigned that Class ID - specifically, get their ID number
- get any students that are assigned that Class ID - specifically, get their ID number
- append the results from teachers and students so that there is a list of all individuals in the class
- look through the “individual data” table to find any individuals with matching ID numbers, and keep those individuals’ birth days.
It is helpful to develop the ability to lay out a set of tables in a schema (because often, database schemas aren’t well documented) and mentally map out the steps that you need to combine tables to get the information you want from the information you have.
Table joins allow us to combine information stored in different tables, keeping certain information (the stuff we need) while discarding extraneous information.
keys are values that are found in multiple tables that can be used to connect the tables. A key (or set of keys) uniquely identify an observation. A primary key identifies an observation in its own table. A foreign key identifies an observation in another table.
There are 3 main types of table joins:
Mutating joins, which add columns from one table to matching rows in another table
Ex: adding birthday to the table of all individuals in a classFiltering joins, which remove rows from a table based on whether or not there is a matching row in another table (but the columns in the original table don’t change)
Ex: finding all teachers or students who have class ClassIDSet operations, which treat observations as set elements (e.g. union, intersection, etc.)
Ex: taking the union of all student and teacher IDs to get a list of individual IDs
4.5.1 Animating Joins
Note: all of these animations are stolen from https://github.com/gadenbuie/tidyexplain.
If we start with two tables, x and y,
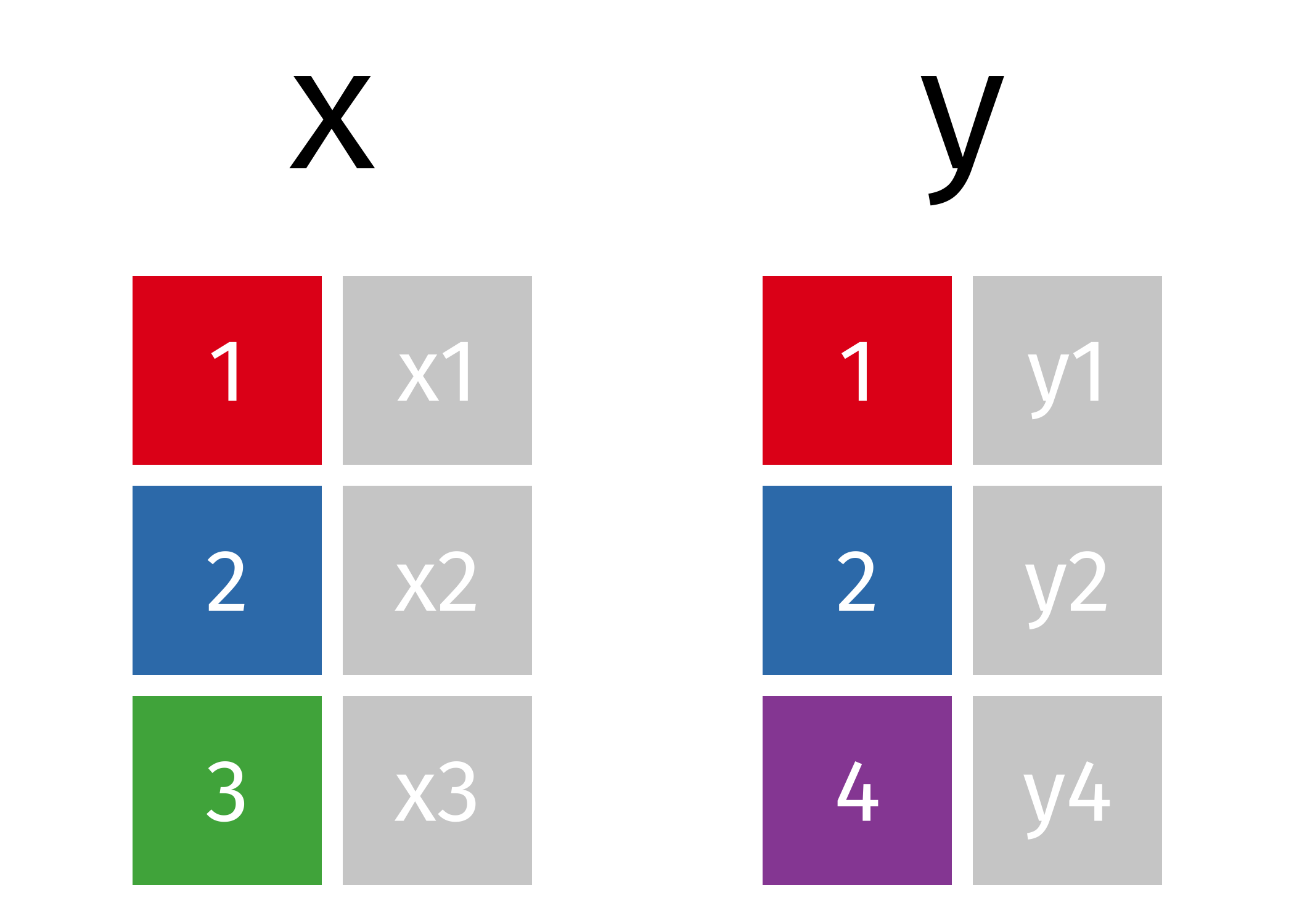
Mutating Joins
We’re primarily going to focus on mutating joins, as filtering joins can be accomplished by … filtering … rather than by table joins.
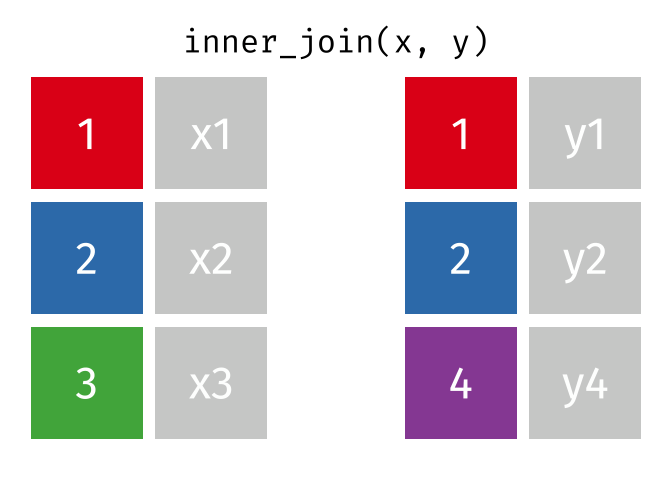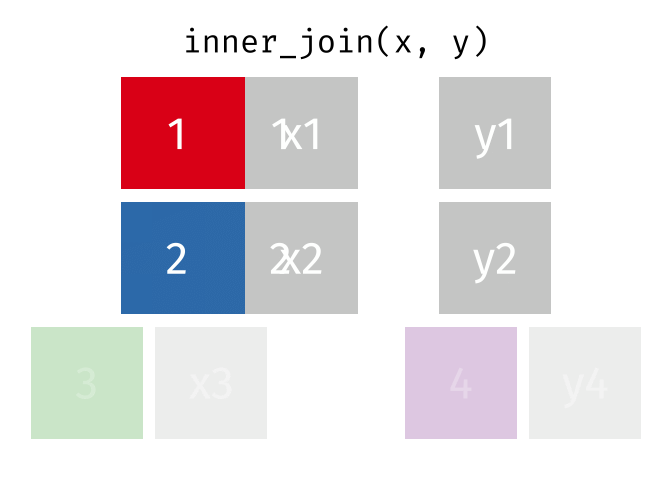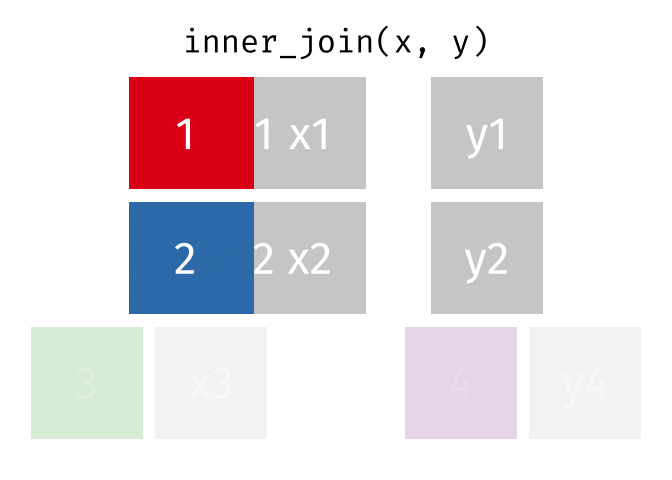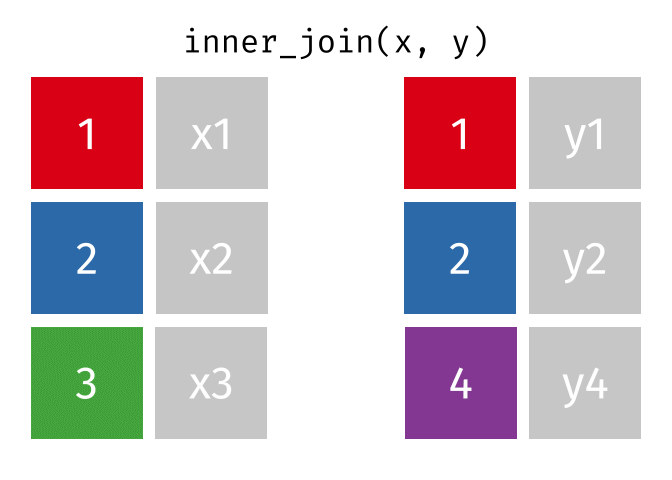
We can do a filtering inner_join to keep only rows which are in both tables (but we keep all columns)
But what if we want to keep all of the rows in x? We would do a left_join
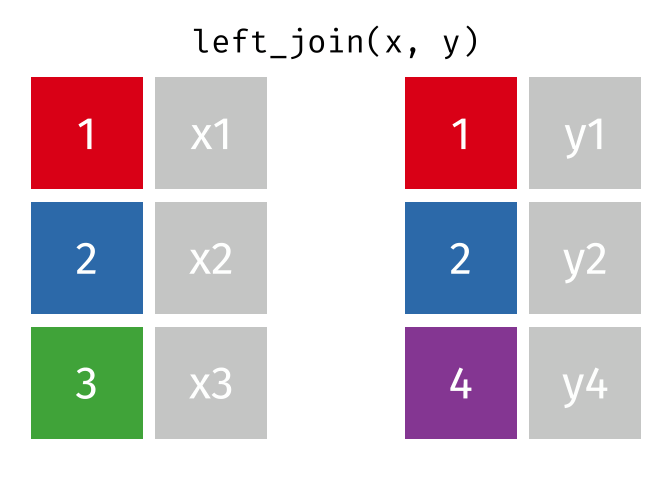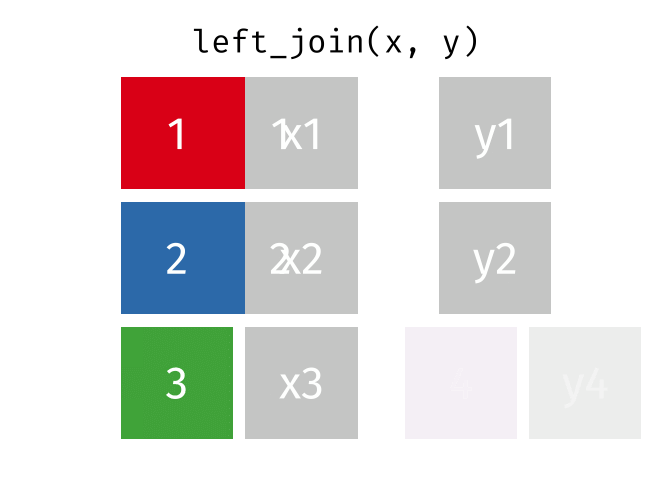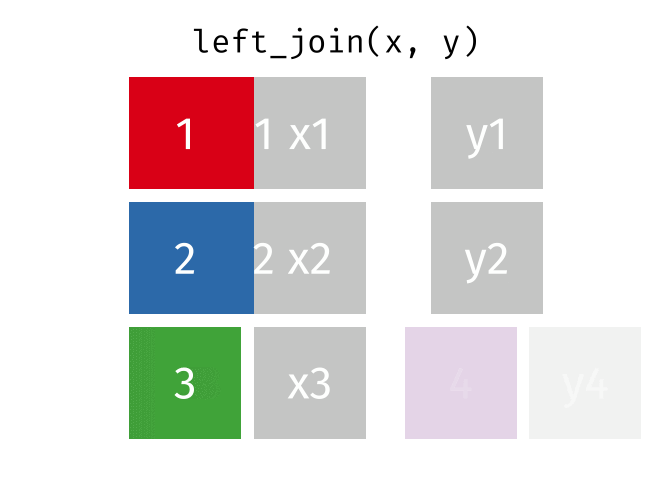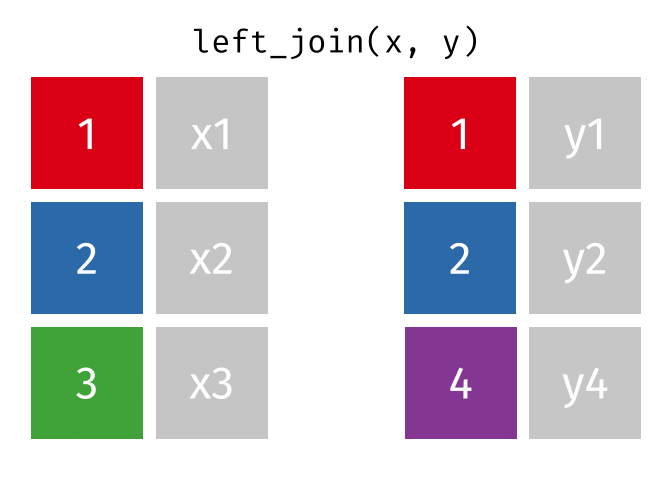
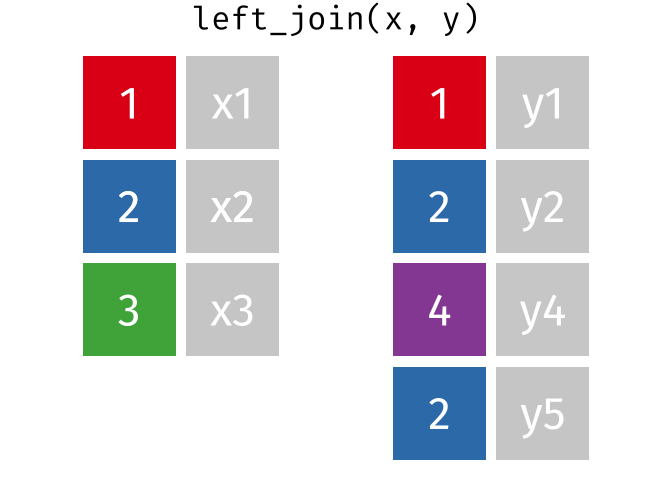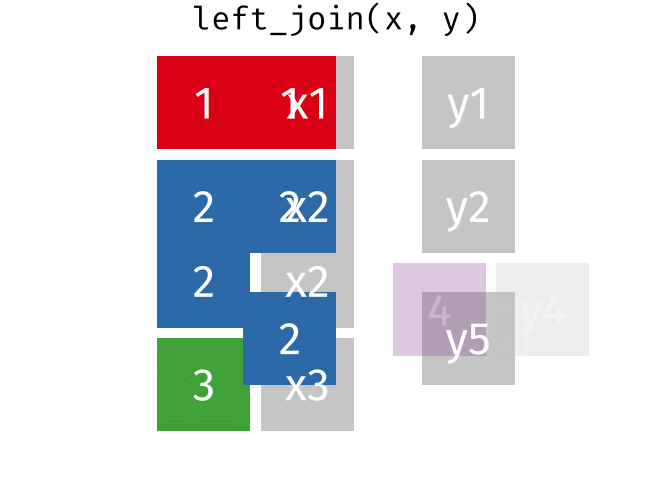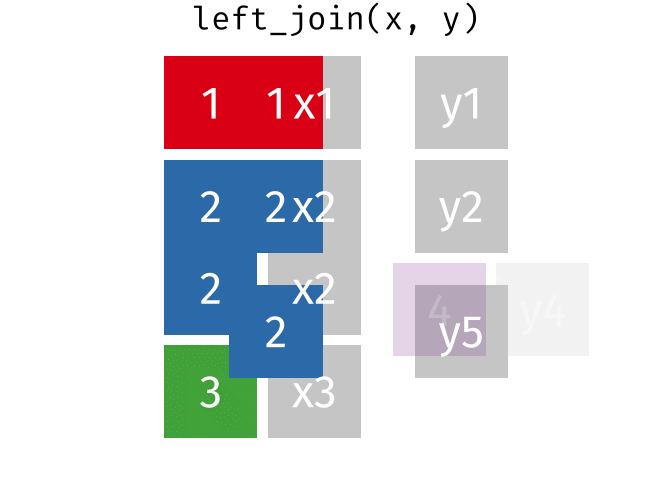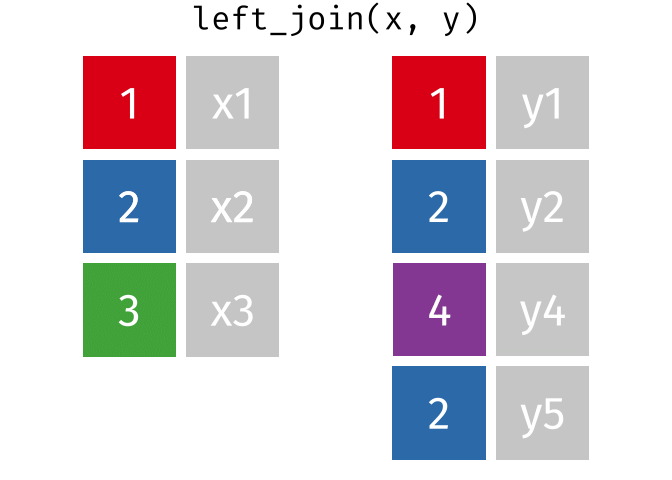
If there are multiple matches in the y table, though, we might have to duplicate rows in x. This is still a left join, just a more complicated one.
If we wanted to keep all of the rows in y, we would do a right_join:
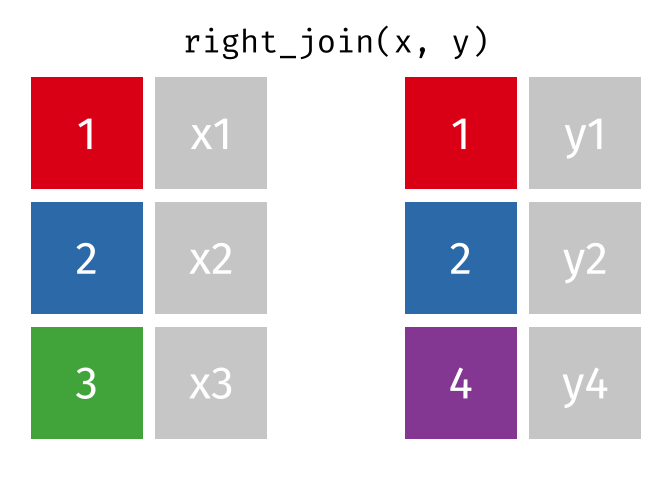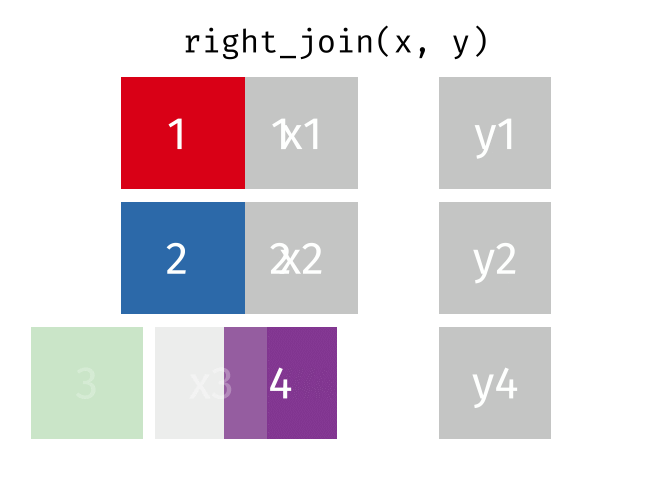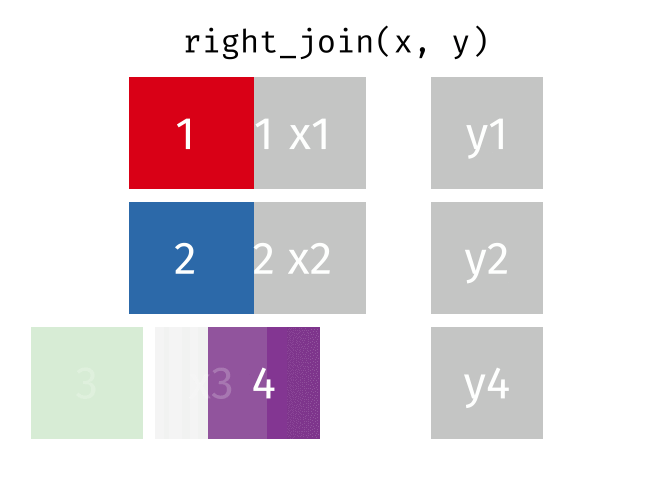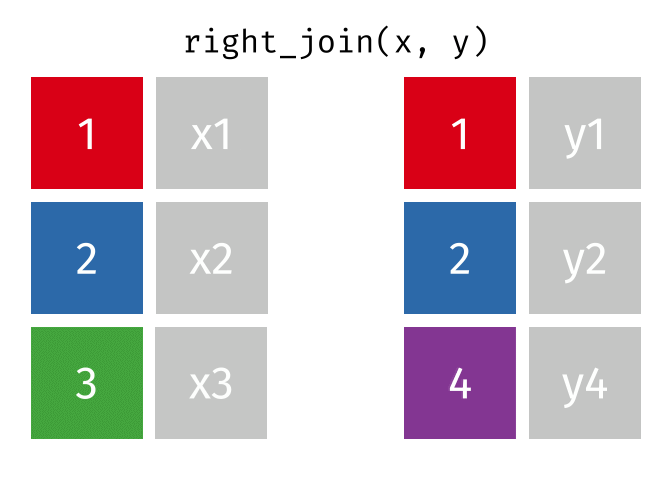
(or, we could do a left join with y and x, but… either way is fine).
And finally, if we want to keep all of the rows, we’d do a full_join:
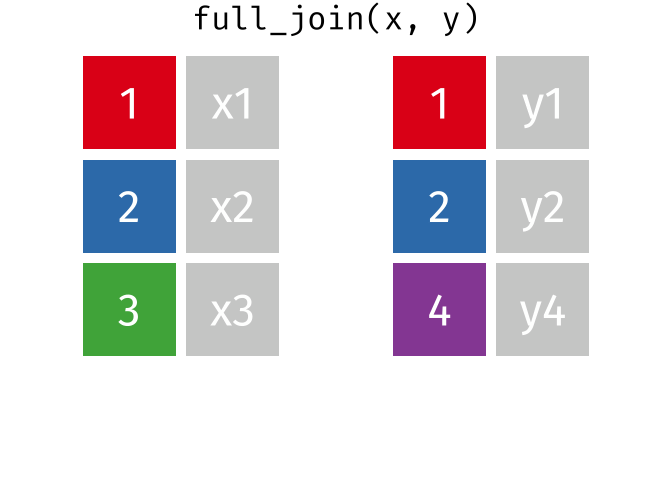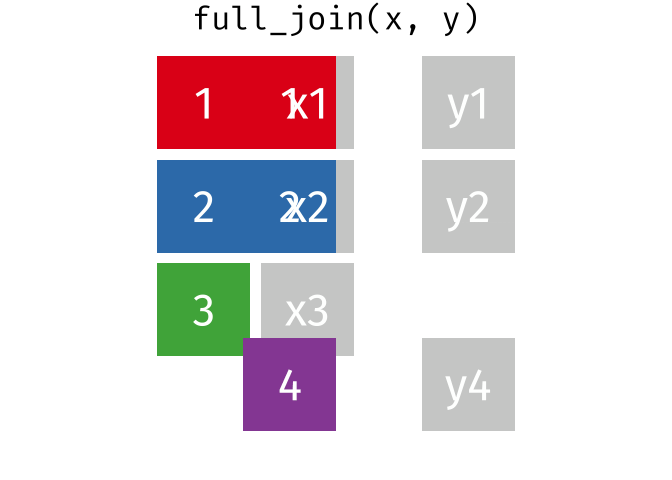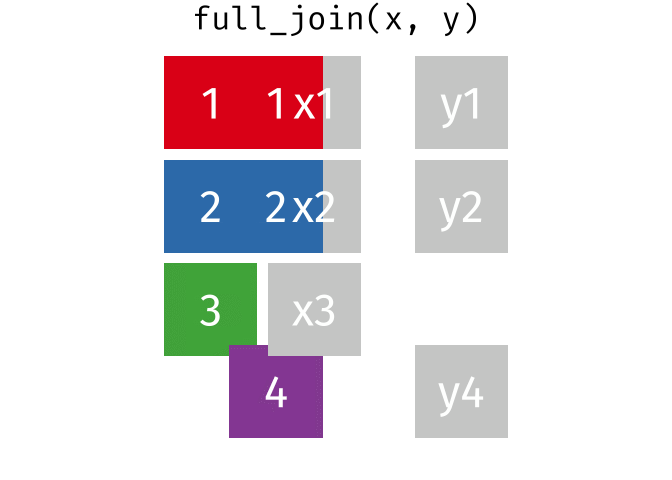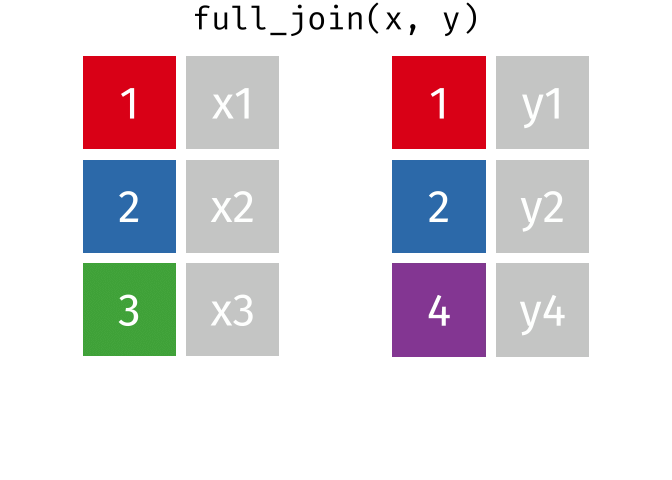
You can find other animations corresponding to filtering joins and set operations here
Every join has a “left side” and a “right side” - so in some_join(A, B), A is the left side, B is the right side.
Joins are differentiated based on how they treat the rows and columns of each side. In mutating joins, the columns from both sides are always kept.
| Left Side | Right Side | ||
| Join Type | Rows | Cols | |
| inner | matching | all | matching |
| left | all | all | matching |
| right | matching | all | all |
| outer | all | all | all |
Demonstration: Mutating Joins
# A tibble: 3 × 2
x y
<chr> <dbl>
1 A 1
2 B 2
3 D 3# A tibble: 3 × 2
x z
<chr> <dbl>
1 B 2
2 C 4
3 D 5An inner join keeps only rows that exist on both sides, but keeps all columns.
inner_join(t1, t2)# A tibble: 2 × 3
x y z
<chr> <dbl> <dbl>
1 B 2 2
2 D 3 5A left join keeps all of the rows in the left side, and adds any columns from the right side that match rows on the left. Rows on the left that don’t match get filled in with NAs.
left_join(t1, t2)# A tibble: 3 × 3
x y z
<chr> <dbl> <dbl>
1 A 1 NA
2 B 2 2
3 D 3 5left_join(t2, t1)# A tibble: 3 × 3
x z y
<chr> <dbl> <dbl>
1 B 2 2
2 C 4 NA
3 D 5 3There is a similar construct called a right join that is equivalent to flipping the arguments in a left join. The row and column ordering may be different, but all of the same values will be there
right_join(t1, t2)# A tibble: 3 × 3
x y z
<chr> <dbl> <dbl>
1 B 2 2
2 D 3 5
3 C NA 4right_join(t2, t1)# A tibble: 3 × 3
x z y
<chr> <dbl> <dbl>
1 B 2 2
2 D 5 3
3 A NA 1An outer join keeps everything - all rows, all columns. In dplyr, it’s known as a full_join.
full_join(t1, t2)# A tibble: 4 × 3
x y z
<chr> <dbl> <dbl>
1 A 1 NA
2 B 2 2
3 D 3 5
4 C NA 4Filtering Joins
A semi join keeps matching rows from x and y, discarding all other rows and keeping only the columns from x.

An anti-join keeps rows in x that do not have a match in y, and only keeps columns in x.

Learn More in R4DS
Read more about joins in r4ds